Scheme Extensions
|
|
|
Technical
Article |
| Scheme is a public domain programming
language, based on the LISP language, that uses an interpreter to run commands. ACIS
provides extensions (written in C++) to the native
Scheme language that can be used by an application to interact with ACIS through its
Scheme Interpreter. The C++ source files for ACIS
Scheme extensions are provided with the product. Spatial's Scheme based demonstration
application, Scheme ACIS Interface Driver Extension (Scheme AIDE), also uses these Scheme
extensions and the Scheme Interpreter.
|
hh:analyze-body
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_geombuild_check,
api_hh_init_body_for_healing
Syntax- (hh:analyze-body body [acis-opts])
Arg Types-
Returns- real
Description
This extension checks the input body for errors. The tests include all the tests
performed by the individual analyze Scheme extensions for the various specific types of
entities (for example, hh:analyze-coedge).
The results are attached to any bad entities as attributes.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-body
; Load a file containing a bad part
(define load (part:load "heal1.sat"))
;; load
; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define init (hh:init-body-for-healing body1))
;; init
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92 |
[Top]
hh:analyze-coedge
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_coedges
Syntax- (hh:analyze-coedge body [acis-opts])
Arg Types-
Returns- real
- Description
This extension checks all the coedges of the input body. The results are attached to any
bad coedges as attributes. The tests include:
Does the coedge lie on the corresponding face surface?- If the coedge contains a pcurve, does the domain of the pcurve correspond with the edge?
- Does the coedge have a partner?
- If the coedge contains a pcurve, is the pcurve within tolerance of the edge?
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-coedge
; Load a file containing a bad part
(define load (part:load "heal1.sat"))
;; load
; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define heal (hh:init-body-for-healing body1))
;; heal
(hh:analyze-coedge body1)
;; () |
[Top]
hh:analyze-edge
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_edges
Syntax- (hh:analyze-edge body [acis-opts])
Arg Types-
Returns- real
- Description
This extension checks all the edges of the input body. The results are attached to any
bad edges as attributes. The tests include:
Check curve geometry- Determine convexity
- Check length
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-edge
; Load a file containing a bad part
(define load (part:load "heal1.sat"))
;; load
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing.
(define heal (hh:init-body-for-healing body1))
;; heal
(hh:analyze-edge body1)
;; () |
[Top]
hh:analyze-face
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_faces
Syntax- (hh:analyze-face body [acis-opts])
Arg Types-
Returns- real
- Description
This extension checks all the faces of the input body. The results are attached to any
bad faces as attributes. The tests include:
Check loops- Check surface
- Check face area
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-face
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1]
(hh:analyze-face body1)
;; () |
[Top]
hh:analyze-geom
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_geombuild_check
Syntax- (hh:analyze-geom body [acis-opts])
Arg Types-
Returns- real
Description
This extension performs the analyze stage of the geometry building phase. The geometry
building phase performs all the geometry related healing operations, including fixing of
edge geometries by intersections, snapping surfaces for fixing tangencies, and refitting
spline surfaces.
A check is run on the body and the inaccurate geometries are marked with attributes. An
invalid edge is one in which the edge curve does not lie on the underlying surfaces to
ACIS tolerance. A vertex is marked bad if it does not lie on the edges or faces which are
incident on it. A bad coedge is one whose pcurve and edge curve do not match.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- The entity must be a body. Individual sheet bodies or faces can be combined into
a single body using the hh:combine extension.
; hh:analyze-geom
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-geom body1)
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.01
; analytic solver tol = 0.01
; isospline solver tol = 0.01
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;; () |
[Top]
hh:analyze-loop
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_loops
Syntax- (hh:analyze-loop body [acis-opts])
Arg Types-
Returns- unspecified
- Description
This extension checks all the loops of the input body. The results are attached to any
bad loops as attributes. The tests include:
Is the loop closed?- Checks loop orientation
- Checks loop coedges for gaps.
- Does the loop self-intersect?
- Checks coedges for correct parameter range.
- Verifies the coedges lie on the face surface
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-loop
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1]
(hh:analyze-loop body1)
;; () |
[Top]
hh:analyze-lump
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_lumps
Syntax- (hh:analyze-lump body [acis-opts])
Arg Types-
Returns- unspecified
- Description
This extension checks all the lumps of the input body. The results are attached to any
bad lumps as attributes. The tests include:
- Checks shells for closure
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-lump
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1]
(hh:analyze-lump body1)
;; () |
[Top]
hh:analyze-shell
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_shells
Syntax- (hh:analyze-shell body [acis-opts])
Arg Types-
Returns- unspecified
- Description
This extension checks all the shells of the input body. The results are attached to any
bad shells as attributes. The tests include:
Checks that the shell is closed- Checks shell orientation
- Checks if shell represents a single volume
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-shell
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1]
(hh:analyze-shell body1)
;; () |
[Top]
hh:analyze-simplify
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_simplify_analyze
Syntax- (hh:analyze-simplify body [simp-opts] [acis-opts])
Arg Types-
Returns- unspecified
Description
This extension analyzes the body
and intelligently sets values of required options and tolerances for geometry simplification.
Geometry simplification attempts to simplify NURBS surfaces into analytic forms (planes,
cylinders, cones, tori, and spheres). If the body is fully analytic, this extension sets a
flag in the simplification aggregate attribute indicating that no geometry simplification is needed.
- Arguments
- body specifies the input body.
simp-opts specifies the optional argument, Simp_Options
object that has been added since ACIS version R11. It sets the parameters which govern the
functionality of hh:analyze-simplify. The constructor of Simp_Options sets the values
of all the parameters as -1. The parameters that can be set are:-
simplify_pos_tol has a default value of -1, that is the scheme command
computes a default value based on its own heuristics.-
max_radius has a default value of -1, that is, a maximum radius of 10,000.
-
do_curve_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1,
that is, curve simplication behavior is set to ON.
-
do_surface_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1,
that is, surface simplication behavior is set to ON.
-
- Note:
(1) Selecting -1 as the value for any of these parameters enables the respective algorithm
to employ its default behavior.
(2) In ACIS R11 and later, both curve simplification and surface simplification are done
by default. In R10 and earlier, only surface simplification was done by default and
there was no curve simplification. To make hh:analyze-simplify behave as in R10
or an earlier release, the caller of the API requires creating a simp_options options object.
Do this by setting do_curve_simplification to "0" and passing this object into the scheme command.
acis-opts is an optional argument enabling journaling and versioning options.
Limitations- The entity must be a body. Individual sheet bodies or faces can be combined into
a single body using the hh:combine extension.
; hh:analyze-simplify
; Analyze a body for possible simplification.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-simplify body1)
; SIMPLIFICATION ANALYSIS :
; =========================
; INPUT STATISTICS :
; 4 Splines,
; 0 Planes,
; 0 Spheres,
; 0 Cylinders,
; 0 Cones,
; 0 Tori
; Simplification not recommended
;; () |
[Top]
hh:analyze-stitch
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_stitch_analyze
Syntax- (hh:analyze-stitch body [acis-opts])
Arg Types-
Returns- unspecified
Description
This extension analyzes the body and intelligently sets values of required options and
tolerances for stitching. Stitching attempts to pair up edges of free faces and stitch
them together. If no stitching is needed, this extension sets a flag in the stitching
aggregate attribute indicating this.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- Entity must be a body. Individual sheet bodies or faces can be combined into a
single body using the hh:combine extension.
; hh:analyze-stitch
; Analyze a body for stitching tolerance.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-stitch body1)
; STITCH ANALYSIS :
; =================
; Input Body Statistics :
; 0 Solids
; 0 Sheets
; 4 Free faces
; Min. Stitch tolerance = 1e-005
; Max. Stitch tolerance = 1
;; () |
[Top]
hh:analyze-vertex
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_analyze_vertices
Syntax- (hh:analyze-vertex entity [acis-opts])
Arg Types-
Returns- unspecified
- Description
This extension checks all the vertices of the input entity(body).
The results are attached to any bad vertices as attributes. The tests include:
Does the vertex lie on the corresponding edges?- Do the edges meet at the vertex?
- Does the vertex lie on the corresponding surfaces?
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:analyze-vertex
; Check vertices for errors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the entities.
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1]
(hh:analyze-vertex body1)
;; () |
[Top]
hh:autoheal
- Action
- Heals anomalies in the input body.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- api_hh_auto_heal
Syntax- (hh:autoheal body [filename] [acis-opts])
Arg Types-
Returns- unspecified
Description
This extension takes a body and returns a body which may be partially healed. The body
may consist of individual faces built with the hh:combine extension.
The extension scans the input entity looking for inconsistencies in the topology. It
removes some inconsistencies, such as edges without geometry. It also trims the surface
geometries wherever possible.
The steps taken are:
- Preprocess the body, checking for inconsistencies such as edges without geometry. Trim
surface geometries whenever possible.
- Test and simplify any spline surfaces which meet the testing criteria. For example, if
the spline surface can be represented by an analytic surface within the tolerance, replace
the spline surface with the appropriate analytic surface (plane, cylinder, etc.).
- If the input consists of faces, determine a stitching tolerance, and attempt to stitch
the faces into either a complete solid or a single sheet body.
- Analyze and heal the stitched geometry:
Build constraints by analyzing all tangency conditions.
Solve analytic surface constraints by modifying analytic surfaces to satisfy the
tangent constraints.
Re-intersect transverse edges.
Solve tangent spline constraints by modifying the associated spline surfaces.
If the optional filename string is supplied, a summary will be written to the file. A
suffix of .log is appended to the filename.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:autoheal
; Autoheal a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(define zoom (zoom-all))
;; zoom
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:autoheal body1 "log")
; PRE-PROCESS RESULTS :
; =====================
; No of zero edges removed : 0
; No of zero faces removed : 0
; No of duplicate vertices : 0
; No of edge grouping : 0
;
; SIMPLIFICATION ANALYSIS :
; =========================
; INPUT STATISTICS :
; 4 Splines, 0 Planes, 0 Spheres, 0 Cylinders,
; 0 Cones, 0 Tori present in the body
; Simplification not recommended
;
; SIMPLIFICATION FIX RESULTS :
; ============================
;
; Simplification tolerance = 0.0001
; no. of initial splines = 4
; no. of final splines = 4
; 0 Planes, 0 Spheres, 0 Cylinders, 0 Cones,
; 0 Tori got made from splines
;
; STITCH ANALYSIS :
; =================
;
; Input Body Statistics :
; 0 Solids
; 0 Sheets
; 4 Free faces
;
; Min. Stitch tolerance = 1e-05
; Max. Stitch tolerance = 1
;
; Stitching 4 lumps at tolerence 0.00001
; Stitching 3 lumps at tolerence 0.000025
; Stitching 3 lumps at tolerence 0.00005
; Stitching 3 lumps at tolerence 0.000075
; Stitching 3 lumps at tolerence 0.0001
; Stitching 3 lumps at tolerence 0.00025
; Stitching 3 lumps at tolerence 0.0005
; Stitching 3 lumps at tolerence 0.00075
; Stitching 3 lumps at tolerence 0.001
; Stitching 3 lumps at tolerence 0.0025
; Stitching 3 lumps at tolerence 0.005
; Stitching 3 lumps at tolerence 0.0075
; Stitching 3 lumps at tolerence 0.01
; Stitching 3 lumps at tolerence 0.025
;
; STITCH CALCULATION RESULTS :
; ============================
; min tol used = 1e-05
; max tol used = 1
; no. solid lumps expected = 0
; no. sheet lumps expected = 1
; no. free faces expected = 0
; no. unshared loops expected = 1
; no. unshared edges expected = 9
;
; STITCH RESULTS :
; ================
; min_tol = 1e-05
; max_tol = 1
; no. solid lumps made = 0
; no. sheet lumps made = 1
; no. free faces remaining = 0
; no. unshared loops = 1
; no. unshared edges = 9
;
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.075
; analytic solver tol = 0.01
; isospline solver tol = 0.075
; no. of edges = 13
; no. of bad edges = 7
; no. of coedges = 17
; no. of bad coedges = 17
; no. of vertices = 10
; no. of bad vertices = 2
; no. of bad tangent edges = 2
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 1
; no. of bad tangent edges boundary uv_uv = 1
; no. of bad tangent edges uv_nonuv = 1
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 1
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 37
;
; ANALYTIC SOLVER RESULTS :
; =========================
; 1 degree of snapper graph
; 0 analytic tangent junctions resolved
; 0 analytic tangent junctions unresolved
; 0 analytic intersections resolved
; 0 analytic intersections unresolved
; 0 vertices resolved (0 intersected,
; 0 projected)
; 0 unstable vertices corrected
; 0 vertices unresolved
; 0 edges calculated by exact projections
; 0 edges calculated by approx projections
; 0 coincident snaps resolved
;
;
; ISOSPLINE SOLVER RESULTS :
; ==========================
; 1 isospline tangent junctions resolved
; 0 isospline tangent junctions unresolved
; 0 splines bent to vertices
;
; SHARP EDGE SOLVER RESULTS :
; ===========================
; 5 sharp edges resolved
; (2 intersected, 0 exact projections,
; 3 approx projections)
; 0 sharp edges unresolved
; 2 vertices resolved
; (0 intersected, 2 exact projections,
; 0 approx projections)
; 0 vertices unresolved
;
; GEN-SPLINE RESULTS :
; ====================
; 1 4-sided patches made
; 0 3-sided patches made
; 0 failures
; 0 unsolvable junctions
;
; WRAPUP ANALYSE RESULTS :
; ========================
; 17 Coedges bad.
;
; WRAPUP RESULTS :
; ================
; 17 pcurves computed
; 4 edges trimmed
;
; GEOMBUILD CALCULATION RESULTS :
; ===============================
;
; Analytic Solver :
; 1 degree of snapper graph
; 0 analytic tangent junctions resolved
; 0 analytic tangent junctions unresolved
; 0 analytic intersections resolved
; 0 analytic intersections unresolved
; 0 vertices resolved (0 intersected,
; 0 projected)
; 0 unstable vertices corrected
; 0 vertices unresolved
; 0 edges calculated by exact projections
; 0 edges calculated by approx projections
; 0 coincident snaps resolved
;
; Isospline Solver :
; 1 isospline tangent junctions resolved
; 0 isospline tangent junctions unresolved
; 0 splines bent to vertices
;
; Reblending :
; 0 faces classified as blends
;
; Sharp Edge Solver :
; 5 sharp edges resolved
; (2 intersected, 0 exact projections,
; 3 approx projections)
; 0 sharp edges unresolved
; 2 vertices resolved
; (0 intersected, 2 exact projections,
; 0 approx projections)
; 0 vertices unresolved
;
; Generic Spline Solver :
; 1 4-sided patches made
; 0 3-sided patches made
; 0 failures
; 0 unsolvable junctions
;
; Wrapup Module :
; 17 pcurves computed
; 4 edges trimmed
;
; GEOMBUILD FIX RESULTS :
; =======================
; Statistics of the healed body after
; geombuild fix :
; no. of edges = 13
; no. of bad edges = 0
; no. of coedges = 17
; no. of bad coedges = 0
; no. of vertices = 10
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 100
;
; Post-processing body after healing ...
; *** Warning (kernel/sg_husk/net:BAD_SPLINE_APPROX)
; Bad spline approximation
;; (#[entity 6 1] 72 100)
; Remove healing attributes
(hh:terminate-body-for-healing body1)
;; #[entity 6 1] |
[Top]
hh:combine
- Action
- Combines a list of solid and/or wire bodies into a single body.
Filename- scm/scmext/heal/scm_heal.cpp
APIs-
api_mk_by_faces
Syntax- (hh:combine body-list [acis-opts])
Arg Types-
Returns- body
- Arguments
- body-list specifies the list of two or more sheet bodies or faces to be combined. This
extension combines all sheet bodies or faces into one body.The resulting body assumes the
entity number of the first body in the list if the input contains sheet bodies. If the
list contains faces, a new body is created.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:combine
; Combine two blocks into a single body.
(define block1
(solid:block (position 0 0 0)
(position 10 20 20)))
;; block1
; Create block 2.
(define block2
(solid:block (position 20 0 0)
(position 30 40 40)))
;; block2
; OUTPUT Original
; Combine the blocks into a single body
; with two lumps.
(define body1 (hh:combine (list block1 block2)))
;; body1
; OUTPUT Result
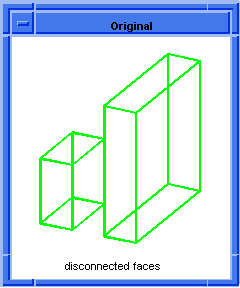  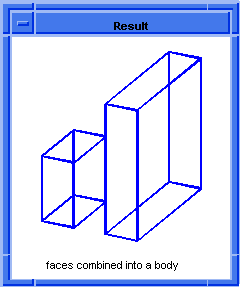
Figure. hh:combine |
[Top]
hh:geombuild
- Action
- Automatically executes all the analyze, calculate, and fix stages of the geometry
building phase.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_geombuild_auto
Syntax- (hh:geombuild body [acis-opts])
Arg Types-
Returns- body
Description
This extension automatically performs the analyze, calculate, and fix stages of the
geometry building phase. Intelligent tolerances that are recommended by the analyze stage
are used in the calculate stage. The geometry building phase performs all the geometry
related healing operations, including fixing of edge geometries by intersections, snapping
surfaces for fixing tangencies, and refitting spline surfaces.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- Entity must be a body. Individual sheet bodies or faces can be combined into a
single body using the hh:combine extension.
; hh:geombuild
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Output the results of geombuild to a file
(debug:file "geom.log")
;; #[file-input-output-port "geom.log"]
(define geom (hh:geombuild body1))
;; geom
; Close the debug file
(debug:file "stdout")
;; #[file-input-output-port "stdout"]
; Open the text file "geom.log" to see the
; following results of hh:geombuild.
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.01
; analytic solver tol = 0.01
; isospline solver tol = 0.01
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;
; GEOMBUILD CALCULATION RESULTS :
; ===============================
; Analytic Solver :
; 1 degree of snapper graph
; 0 analytic tangent junctions resolved
; 0 analytic tangent junctions unresolved
; 0 analytic intersections resolved
; 0 analytic intersections unresolved
; 0 vertices resolved (0 intersected,
; 0 projected)
; 0 unstable vertices corrected
; 0 vertices unresolved
; 0 edges calculated by exact projections
; 0 edges calculated by approx projections
; 0 coincident snaps resolved
;
; Isospline Solver :
; 0 isospline tangent junctions resolved
; 0 isospline tangent junctions unresolved
; 0 splines bent to vertices
;
; Reblending :
; 0 faces classified as blends
;
; Sharp Edge Solver :
; 3 sharp edges resolved
; (0 intersected, 0 exact projections,
; 3 approx projections)
; 0 sharp edges unresolved
; 0 vertices resolved
; (0 intersected, 0 exact projections,
; 0 approx projections)
; 0 vertices unresolved
;
; Generic Spline Solver :
; 0 4-sided patches made
; 0 3-sided patches made
; 0 failures
; 0 unsolvable junctions
;
; Wrapup Module :
; 4 pcurves computed
; 0 edges trimmed
;
; GEOMBUILD FIX RESULTS :
; =======================
; Statistics of the healed body after
; geombuild fix :
; no. of edges = 17
; no. of bad edges = 0
; no. of coedges = 17
; no. of bad coedges = 0
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 100 |
[Top]
hh:geombuild-analyze
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_geombuild_analyze
Syntax- (hh:geombuild-analyze body [acis-opts])
Arg Types-
Returns- body
Description
This extension automatically performs the analyze, calculate, and fix stages of the
geometry building phase. Intelligent tolerances that are recommended by the analyze stage
are used in the calculate stage. The geometry building phase performs all the geometry
related healing operations, including fixing of edge geometries by intersections, snapping
surfaces for fixing tangencies, and refitting spline surfaces.
- Arguments
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- Entity must be a body. Individual sheet bodies or faces can be combined into a
single body using the hh:combine extension.
; hh:geombuild-analyze
; Load a file containing a bad part
(define load (part:load "heal1.sat"))
;; load
; Zoom the view in order to see the part.
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body.
(define body1 (hh:combine (part:entities)))
;; body1
; Heal the body.
(define heal (hh:geombuild-analyze body1))
;; heal |
[Top]
hh:get-analytic-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-analytic-tol body)
Arg Types-
Returns- real
Description
The analytic tolerance is used in the analytic solver subphase of geometry building,
which fixes tangency constraints in the model. This is an upper bound for deviation of the
analytic surfaces to satisfy the constraints. The default value of 0.01 permits
translations of 0.01 to be performed to surfaces.
- Arguments
- Input argument is the body whose analytic solver tolerance is to be obtained.
; hh:get-analytic-tol
; Get the analytic healing tolerance for a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do geombuild analysis
(hh:analyze-geom body1)
;; ()
; Get the analytic tolerance
(hh:get-analytic-tol body1)
;; 0 |
[Top]
hh:get-geombuild-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-geombuild-tol body)
Arg Types-
Returns- real
Description
The geometry build tolerance drives the actual geometry building of the model. This
should typically be slightly more (around 3 times) than the maximum gap size in the model.
The maximum gap size calculated during stitching is used as the geometry building
tolerance for automatic healing (autoheal). However, the user may need to increase the
geometry building tolerance if the healed geometry deviates substantially from the
original geometry.
- Arguments
- Input argument is the body whose geometric build solver tolerance is to be obtained.
; hh:get-geombuild-tol
; Get the geometry build healing tolerance
; for a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do geombuild analysis
(hh:analyze-geom body1)
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.01
; analytic solver tol = 0.01
; isospline solver tol = 0.01
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;; ()
; Now get tolerance
(hh:get-geombuild-tol body1)
;; 100 |
[Top]
hh:get-isospline-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-isospline-tol body)
Arg Types-
Returns- unspecified
Description
The isospline solver attempts to heal all edges shared by tangential isoparametric
surfaces (for example, the intersection curve is an isoparametric curve of both splines in the
intersection). It calculates isoparametric junctions of spline geometries intersecting
tangentially.
- Arguments
- Input argument is the body whose isospline solver tolerance is to be obtained.
; hh:get-isospline-tol
; Get the isospline healing tolerance for a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do geombuild analysis
(hh:analyze-geom body1)
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.01
; analytic solver tol = 0.01
; isospline solver tol = 0.01
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;; ()
; Now get tolerance
(hh:get-isospline-tol body1)
;; 0 |
[Top]
hh:get-simplify-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-simplify-tol body)
Arg Types-
Returns- real
Description
The geometry simplification tolerance is the tolerance at which spline surfaces get
simplified to analytic surfaces. If the tolerance is tight (as default is), only spline
surfaces that are exact analytic surfaces get simplified. If the tolerance is loosened,
then approximate analytic fits to splines are obtained. In such cases, the gaps between
surfaces may increase and healing in subsequent operations may be more difficult. The need
for increasing tolerances typically arises when analytic surfaces are output as NUBS
surfaces rather than NURBS surfaces. The default tolerance is 0.0001 (length units), which
obtains a very good approximation of analytic surfaces to spline surfaces in most models.
- Arguments
- Input argument is the body whose geometry simplification tolerance is to be obtained for
healing.
; hh:get-simplify-tol
; Get the simplify healing tolerance for a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do simplify analysis
(hh:analyze-simplify body1)
; SIMPLIFICATION ANALYSIS :
; =========================
; INPUT STATISTICS :
; 4 Splines, 0 Planes, 0 Spheres, 0 Cylinders,
; 0 Cones, 0 Tori present in the body
; Simplification not recommended
;; ()
; Get tolerance
(hh:get-simplify-tol body1)
;; 0.0001 |
[Top]
hh:get-stitch-max-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-stitch-max-tol body)
Arg Types-
Returns- real
Description
The minimum and maximum stitching tolerances specify the range in which stitching
between edges is performed. The stitching begins from the minimum tolerance and increases
in steps towards the maximum tolerance.
- Arguments
- Input argument is the body whose maximum stitching tolerance is to be obtained for
healing.
; hh:get-stitch-max-tol
; Get the maximum stitching healing tolerance
; for a body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do stitch analysis
(hh:analyze-stitch body1)
; STITCH ANALYSIS :
; =================
; Input Body Statistics :
; 0 Solids
; 0 Sheets
; 4 Free faces
; Min. Stitch tolerance = 1e-005
; Max. Stitch tolerance = 1
;; ()
; Get tolerance
(hh:get-stitch-max-tol body1)
;; 1 |
[Top]
hh:get-stitch-min-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:get-stitch-min-tol body)
Arg Types-
Returns- real
Description
The minimum and maximum stitching tolerances specify the range in which stitching
between edges is performed. The stitching begins from the minimum tolerance and increases
in steps towards the maximum tolerance.
- Arguments
- Input argument is the body whose minimum stitching tolerance is to be obtained for
stitching.
; hh:get-stitch-min-tol
; Get the minimum stitching tolerance.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Do stitch analysis
(hh:analyze-stitch body1)
; STITCH ANALYSIS :
; =================
; Input Body Statistics :
; 0 Solids
; 0 Sheets
; 4 Free faces
; Min. Stitch tolerance = 1e-005
; Max. Stitch tolerance = 1
;; ()
; Get tolerance
(hh:get-stitch-min-tol body1)
;; 1e-05 |
[Top]
hh:init-body-for-healing
- Action
- Removes any existing healing attributes and attaches new healing attributes.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_init_body_for_healing
Syntax- (hh:init-body-for-healing body)
Arg Types-
Returns- body
Description
This extension removes any previously defined healing attributes and then attaches a
new set of healing attributes to the body.
This extension should be called before any other healing extension.
; hh:init-body-for-healing
; Attach healing attributes
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:init-body-for-healing body1)
;; #[entity 6 1] |
[Top]
hh:postprocess
- Action
- Cleans the healed model.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_postprocess
Syntax- (hh:postprocess body [acis-opts])
Arg Types-
Returns- body
Description
This extension performs post-processing of the healed data. It performs such operations
as correction of negative area faces, duplicate vertices, and edge groups.
This extension should be called as the final step when healing is accomplished using
individual healing components instead of autohealing.
- Arguments
- Input argument is the body that has been healed.
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- The entity must be a body or a sheet body.
; hh:postprocess
; Postprocess a healed body.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define preprocess (hh:preprocess body1))
;; preprocess
(define process (hh:postprocess body1))
;; process |
[Top]
hh:preprocess
- Action
- Cleans the model to be healed.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_preprocess
Syntax- (hh:preprocess body [acis-opts])
Arg Types-
Returns- body
Description
This extension scans the input entity to locate inconsistencies in the topology. It
removes some inconsistencies, such as zero-length edges, sliver faces, and duplicate
vertices.
This extension should be called as the first step when healing is to be accomplished
using individual healing components instead of autohealing.
- Arguments
- Input argument is the body that has to be healed.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:preprocess
; Preprocess a body for healing.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define preprocess (hh:preprocess body1))
;; preprocess |
[Top]
hh:reset-tols
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:reset-tols)
Arg Types-
Returns- unspecified
Description
Resets all the user define tolerances flags to false so that subsequent healing
operations will use the system defaults for all tolerance values.
; hh:reset-tols
; Set the isospline healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-isospline-tol 0.01)
;; ()
; Autoheal will use user-defined tolerance
(hh:autoheal body1)
; ... (autoheal results)
;; (#[entity 6 1] 72 100)
; Now reset tolerance flags to use default values for
; subsequent healing operations
(hh:reset-tols)
;; () |
[Top]
hh:set-analytic-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-analytic-tol tolerance)
Arg Types-
Returns- unspecified
- Arguments
- The analytic tolerance is used in the analytic solver subphase of geometry building,
which fixes tangency constraints in the model. This is upper bound for deviation of the
analytic surfaces to satisfy the constraints. The default value of 0.01 permits
translations of 0.01 to be performed to the surfaces.
; hh:set-analytic-tol
; Set the analytic healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-analytic-tol 0.01)
;; () |
[Top]
hh:set-geombuild-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-geombuild-tol tolerance)
Arg Types-
Returns- unspecified
Description
The geometry build tolerance drives the actual geometry building of the model. This
should typically be slightly more (around 3 times) than the maximum gap size in the model.
The maximum gap size calculated during stitching is used as the geometry building
tolerance for automatic healing (autoheal). However, the user may need to increase the
geometry building tolerance if the healed geometry deviates substantially from the
original geometry.
; hh:set-geombuild-tol
; Set the geombuild healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-geombuild-tol 0.01)
;; () |
[Top]
hh:set-isospline-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-isospline-tol tolerance)
Arg Types-
Returns- unspecified
Description
The isospline solver attempts to heal all edges shared by tangential isoparametric
surfaces (for example, the intersection curve is an isoparametric curve of both splines in the
intersection). It calculates isoparametric junctions of spline geometries intersecting
tangentially.
; hh:set-isospline-tol
; Set the isospline healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-isospline-tol 0.01)
;; () |
[Top]
hh:set-simplify-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-simplify-tol tolerance)
Arg Types-
Returns- unspecified
Description
The geometry simplification tolerance is the tolerance at which spline surfaces get
simplified to analytic surfaces. If the tolerance is tight (as default is), only spline
surfaces that are exact analytic surfaces get simplified. If the tolerance is loosened,
then approximate analytic fits to splines are obtained. In such cases, the gaps between
surfaces may increase and healing in subsequent operations may be more difficult. The need
for increasing tolerances typically arises when analytic surfaces are output as NUBS
surfaces rather than NURBS surfaces. The default tolerance is 0.0001 (length units), which
obtains a very good approximation of analytic surfaces to spline surfaces in most models.
; hh:set-simplify-tol
; Set the simplify healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-simplify-tol 0.01)
;; () |
[Top]
hh:set-stitch-max-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-stitch-max-tol tolerance)
Arg Types-
Returns- unspecified
Description
The minimum and maximum stitching tolerances specify the range in which stitching
between edges is performed. The stitching begins from the minimum tolerance and increases
in steps towards the maximum tolerance.
- Arguments
- Input argument tolerance specifies the maximum stitch tolerance.
; hh:set-stitch-max-tol
; Set the maximum stitching healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-stitch-max-tol 0.01)
;; () |
[Top]
hh:set-stitch-min-tol
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_ah.cpp
APIs- None
Syntax- (hh:set-stitch-min-tol tolerance)
Arg Types-
Returns- unspecified
Description
The minimum and maximum stitching tolerances specify the range in which stitching
between edges is performed. The stitching begins from the minimum tolerance and increases
in steps towards the maximum tolerance.
- Arguments
- Input argument tolerance specifies the minimum stitch tolerance.
; hh:set-stitch-min-tol
; Set the minimum stitching healing tolerance value.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:set-stitch-min-tol 0.01)
;; () |
[Top]
hh:show-bad-coedges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-bad-coedges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any coedge of the input body that has an analysis attribute
indicating the coedge failed one or more of the analysis checks. This extension assumes
that the entity was already analyzed by the appropriate analyze scheme extension, and
should be called after analysis phase in healing.
; hh:show-bad-coedges
; Display bad coedges in a highlight color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(define zoom (zoom-all))
;; zoom
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define preprocess (hh:preprocess body1))
;; preprocess
; Analyze the geometry of the body
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
; ()
;; 92
(hh:show-bad-coedges body1)
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1] #[entity 10 1])
; OUTPUT Example 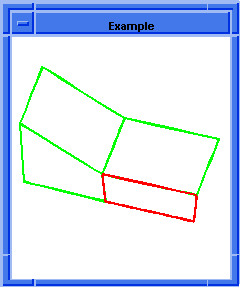
Figure. hh:show-bad-coedges |
[Top]
hh:show-bad-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-bad-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension displays bad edges. Bad edges are those that do not lie on the
underlying surfaces and/or the vertices. This extension should be called after the
geometry analysis phase of healing.
- Arguments
- Input argument is the body whose bad edges are to be highlighted.
; hh:show-bad-edges
; Display bad edges in a highlight color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(define preprocess (hh:preprocess body1))
;; preprocess
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-bad-edges body1)
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1] #[entity 10 1])
; OUTPUT Result
  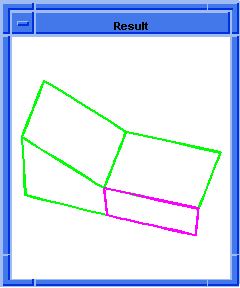
Figure. hh:show-bad-edges |
[Top]
hh:show-bad-faces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-bad-faces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any face of the input body that has an analysis attribute
indicating the face failed one or more of the analysis checks. This extension assumes that
the entity was already analyzed by the appropriate analyze scheme extension. It should be
called after the analysis phase in healing.
; hh:show-bad-faces
; Display bad faces in a highlight color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-bad-faces body1)
;; ()
; OUTPUT Result
  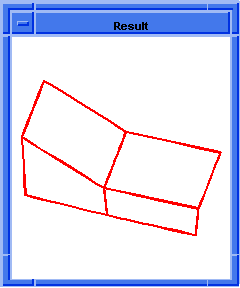
Figure. hh:show-bad-faces |
[Top]
hh:show-bad-vertices
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-bad-vertices body)
Arg Types-
Returns- (entity ...)
Description
This extension displays bad vertices. Bad vertices are those that do not lie on the
underlying surfaces and/or the edges. This extension should be called after the geombuild
analysis phase of healing.
- Arguments
- Input argument is the body whose bad vertices are to be highlighted.
; hh:show-bad-vertices
; Display bad vertices in a highlight color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-bad-vertices body1)
;; () |
[Top]
hh:show-body
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-body body)
Arg Types-
Returns- (entity ...)
- Arguments
- Input argument is the body whose bad geometry is to be highlighted.
; hh:show-body
; Display bad edges, vertices, faces and coedges in
; various highlight colors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-body body1)
; 4 edges bad
; 4 coedges bad
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1]
;; #[entity 10 1] #[entity 11 1] #[entity 12 1]
;; #[entity 13 1] #[entity 14 1])
; OUTPUT Result
  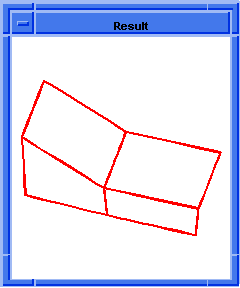
Figure. hh:show-body |
[Top]
hh:show-closed-curves
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-closed-curves body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any curve of the input body that has an analysis attribute
indicating the curve is closed.
; hh:show-closed-curves
; Display closed curves in a highlight color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-closed-curves body1)
;; () |
[Top]
hh:show-closed-surfaces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-closed-surfaces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any surface of the input body that has an analysis attribute
indicating the surface is closed.
; hh:show-closed-surfaces
; Display closed surfaces in a highlight color.
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-closed-surfaces body1)
;; () |
[Top]
hh:show-coedges-no-partner
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-coedges-no-partner body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any coedge of the input body that has an analysis attribute
indicating the coedge does not have a partner.
A list of all coedges in the body that do not have a partner is returned.
; hh:show-coedges-no-partner
; Show coedges without a partner in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-coedges-no-partner body1)
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1]
;; #[entity 10 1] #[entity 11 1] #[entity 12 1]
;; #[entity 13 1] #[entity 14 1] #[entity 15 1]
;; #[entity 16 1] #[entity 17 1] #[entity 18 1]
;; #[entity 19 1] #[entity 20 1] #[entity 21 1]
;; #[entity 22 1] #[entity 23 1])
; OUTPUT Result
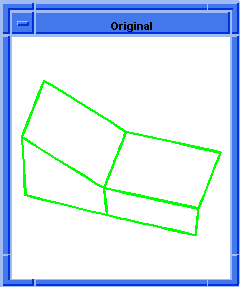  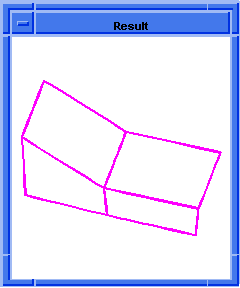
Figure. hh:show-coedges-no-partner |
[Top]
hh:show-coedges-not-on-faces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-coedges-not-on-faces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any coedge of the input body that has an analysis attribute
indicating the coedge does not lie on the surface.
; hh:show-coedges-not-on-faces
; Display coedges not on faces in highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-coedges-not-on-faces body1)
;; (#[entity 7 1] #[entity 8 1])
; OUTPUT Result
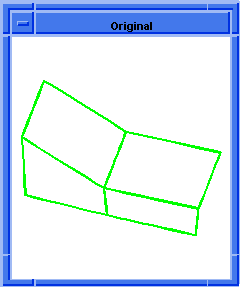  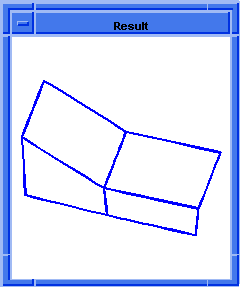
Figure. hh:show-coedges-not-on-faces |
[Top]
hh:show-concave-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-concave-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any edge of the input body that has an analysis attribute
indicating that the edge is concave.
; hh:show-concave-edges
; Display concave edges in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-concave-edges body1)
;; () |
[Top]
hh:show-convex-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-convex-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any edge of the input body that has an analysis attribute
indicating that the edge is convex.
; hh:show-convex-edges
; Display convex edges in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-convex-edges body1)
;; () |
[Top]
hh:show-degenerate-curves
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-degenerate-curves body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any curve of the input body that has an analysis attribute
indicating the curve is degenerate.
; hh:show-degenerate-curves
; Display degenerate curves in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-degenerate-curves body1)
;; () |
[Top]
hh:show-degenerate-surfaces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-degenerate-surfaces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any surface of the input body that has an analysis attribute
indicating the surface is degenerate.
; hh:show-degenerate-surfaces
; Display degenerate surfaces in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-degenerate-surfaces body1)
;; () |
[Top]
hh:show-discontinuous-curves
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-discontinuous-curves body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any curve of the input body that has an analysis attribute
indicating the curve is not continuous.
; hh:show-discontinuous-curves
; Display discontinuous curves in a
; highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-discontinuous-curves body1)
;; () |
[Top]
hh:show-discontinuous-surfaces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-discontinuous-surfaces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any surface of the input body that has an analysis attribute
indicating the surface is not continuous.
; hh:show-discontinuous-surfaces
; Display discontinuous surfaces in a
; highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-discontinuous-surfaces body1)
;; () |
[Top]
hh:show-loops-disoriented
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-loops-disoriented body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any loop of the input body that has an analysis attribute
indicating that the loop has an incorrect orientation.
; hh:show-loops-disoriented
; Display disoriented loops in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-loops-disoriented body1)
;; () |
[Top]
hh:show-loops-gaps
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-loops-gaps body)
Arg Types-
Returns- (entity ...)
- Arguments
- Input argument is the body whose loops that have coedge gaps are to be highlighted.
; hh:show-loops-gaps
; Display loops with gaps in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-loops-gaps body1)
;; () |
[Top]
hh:show-loops-not-on-faces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-loops-not-on-faces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any loop of the input body that has an analysis attribute
indicating that the loop does not lie on the face.
; hh:show-loops-not-on-faces
; Display loops not on faces in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-loops-not-on-faces body1)
;; ()
; OUTPUT Result
  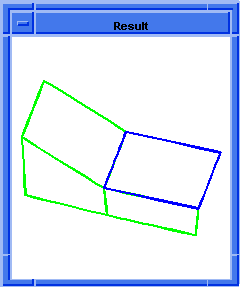
Figure. hh:show-loops-not-on-faces |
[Top]
hh:show-loops-open
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-loops-open body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any loop of the input body that has an analysis attribute
indicating that the loop is open.
; hh:show-loops-open
; Display open loops in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-loops-open body1)
;; () |
[Top]
hh:show-lumps
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_get_bad_lumps
Syntax- (hh:show-lumps body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any lump of the input body that has an analysis attribute
indicating lump is bad.
; hh:show-lumps
; Display lumps in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-lumps body1)
;; ()
; OUTPUT Result
  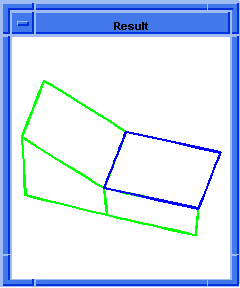
Figure. hh:show-lumps |
[Top]
hh:show-periodic-curves
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-periodic-curves body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any curve of the input body that has an analysis attribute
indicating the curve is periodic.
; hh:show-periodic-curves
; Display periodic curves in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-periodic-curves body1)
;; ()
; OUTPUT Result
  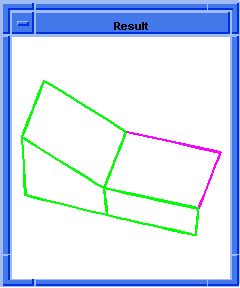
Figure. hh:show-periodic-curves |
[Top]
hh:show-periodic-surfaces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-periodic-surfaces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any surface of the input body that has an analysis attribute
indicating the surface is periodic.
; hh:show-periodic-surfaces
; Display periodic surfaces in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-periodic-surfaces body1)
;; () |
[Top]
hh:show-self-intersecting-curves
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-self-intersecting-curves body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any curve of the input body that has an analysis attribute
indicating the curve is self-intersecting.
; hh:show-self-intersecting-curves
; Display self intersecting surfaces in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-self-intersecting-curves body1)
;; () |
[Top]
hh:show-self-intersecting-surfaces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-self-intersecting-surfaces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any surface of the input body that has an analysis attribute
indicating the surface is self-intersecting.
; hh:show-self-intersecting-surfaces
; Display self intersecting surfaces in a highlighted
; color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-self-intersecting-surfaces body1)
;; () |
[Top]
hh:show-shells
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- api_hh_get_bad_shells
Syntax- (hh:show-shells body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any shell of the input body that has an analysis attribute
indicating that the shell is bad.
; hh:show-shells
; Display shells and bad shells in
; highlighted colors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-shells body1)
;; ()
; OUTPUT Result
  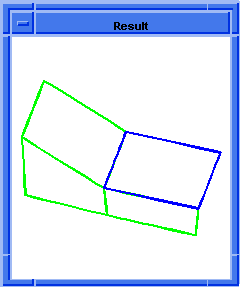
Figure. hh:show-shells |
[Top]
hh:show-short-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-short-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any edge of the input body that has an analysis attribute
indicating that the edge length is less than the geometry tolerance.
; hh:show-short-edges
; Display short edges in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-short-edges body1)
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1]
;; #[entity 10 1] #[entity 11 1] #[entity 12 1]
;; #[entity 13 1] #[entity 14 1] #[entity 15 1]
;; #[entity 16 1] #[entity 17 1] #[entity 18 1]
;; #[entity 19 1] #[entity 20 1] #[entity 21 1]
;; #[entity 22 1] #[entity 23 1])
; OUTPUT Result
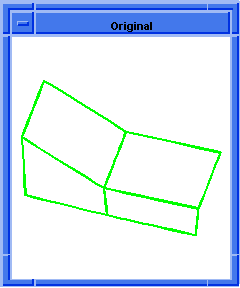  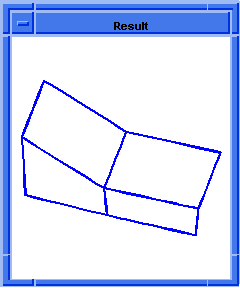
Figure. hh:show-short-edges |
[Top]
hh:show-simplified
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-simplified body)
Arg Types-
Returns- (entity ...)
Description
This extension shows faces that have been simplified. It should be called after the
simplify phase of healing is completed.
- Arguments
- Input argument is the body whose simplified faces are to be highlighted.
; hh:show-simplified
; Display simplified faces in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-simplified body1)
;; () 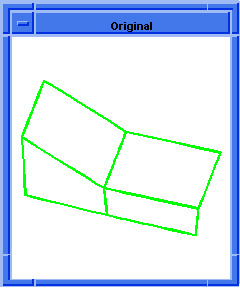  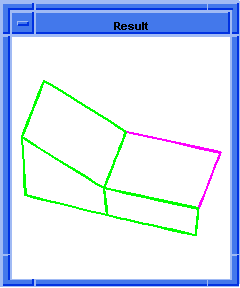
Figure. hh:show-simplified |
[Top]
hh:show-spline
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-spline body)
Arg Types-
Returns- (entity ...)
Description
This extension shows spline faces. It should be called after the simplify analysis is
done.
- Arguments
- Input argument is the body whose spline faces to be simplified are to be highlighted.
; hh:show-spline
; Display spline faces in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-spline body1)
;; () |
[Top]
hh:show-tangent-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-tangent-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any edge of the input body that has an analysis attribute
indicating that the edge joins two tangent faces.
; hh:show-tangent-edges
; Display tangent edges in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT original
(hh:analyze-geom body1)
; GEOMBUILD ANALYSIS :
; ====================
; geom build tol = 0.01
; analytic solver tol = 0.01
; isospline solver tol = 0.01
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;; ()
(hh:show-tangent-edges body1)
;; ()
; OUTPUT Result
  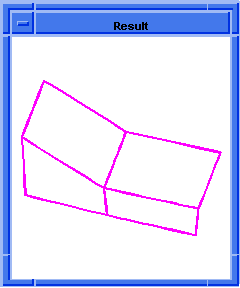
Figure. hh:show-tangent-edges |
[Top]
hh:show-tolerant-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-tolerant-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension changes the color of any edge of the input body that is a tolerant edge.
; hh:show-tolerant-edges
; Display tolerant edges in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Autoheal the body
(hh:autoheal body1)
; ... (autoheal results)
;; (#[entity 6 1] 72 100)
(hh:show-tolerant-edges body1)
;; () |
[Top]
hh:show-unstitched
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_anly.cpp
APIs- None
Syntax- (hh:show-unstitched body)
Arg Types-
Returns- (entity ...)
Description
This extension shows the common edges of unstitched faces. It should be called after
the stitch analysis phase of healing.
- Arguments
- Input argument is the body whose common edges of unstitched faces are to be highlighted.
; hh:show-unstitched
; Display common edges of unstitched faces
; in a highlighted color.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:stitch body1)
; STITCH RESULTS :
; ================
; min_tol = 1e-05
; max_tol = 1
; no. solid lumps made = 0
; no. sheet lumps made = 1
; no. unshared loops = 1
; no. unshared edges = 9
;; #[entity 6 1]
(hh:show-unstitched body1)
;; (#[entity 7 1] #[entity 8 1] #[entity 9 1]
;; #[entity 10 1] #[entity 11 1] #[entity 12 1]
;; #[entity 13 1] #[entity 14 1] #[entity 15 1])
; OUTPUT Result
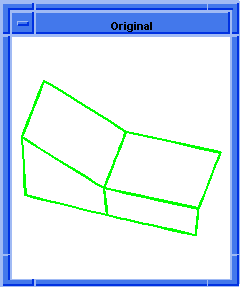  
Figure. hh:show-unstitched |
[Top]
hh:show-vertices-edges-dont-meet
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-vertices-edges-dont-meet body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any vertex of the input body that has an analysis attribute
indicating the vertex is associated with edges that do not meet at the vertex.
; hh:show-vertices-edges-dont-meet
; Display vertices where associated edges
; do not meet in highlighted colors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-vertices-edges-dont-meet body1)
;; ()
; OUTPUT Result
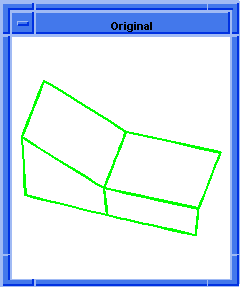  
Figure. hh:show-vertices-edges-dont-meet |
[Top]
hh:show-vertices-not-on-edges
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-vertices-not-on-edges body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any vertex of the input body that has an analysis attribute
indicating the vertex does not lie on the edge.
; hh:show-vertices-not-on-edges
; Display vertices not on edges in highlighted
; colors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-vertices-not-on-edges body1)
;; ()
; OUTPUT Result
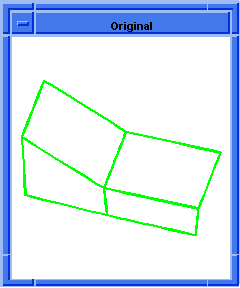  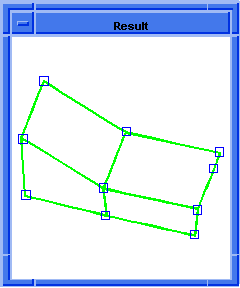
Figure. hh:show-vertices-not-on-edges |
[Top]
hh:show-vertices-not-on-faces
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated direct interface.
Filename- scm/scmext/heal/scm_adv.cpp
APIs- None
Syntax- (hh:show-vertices-not-on-faces body)
Arg Types-
Returns- (entity ...)
Description
This extension highlights any vertex of the input body that has an analysis attribute
indicating the vertex does not lie on the surface.
; hh:show-vertices-not-on-faces
; Display vertices not on faces in highlighted
; colors.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; OUTPUT Original
(hh:analyze-body body1)
; GEOMBUILD CHECK RESULTS :
; Statistics of the body from geombuild check :
; no. of edges = 17
; no. of bad edges = 4
; no. of coedges = 17
; no. of bad coedges = 4
; no. of vertices = 17
; no. of bad vertices = 0
; no. of bad tangent edges = 0
; no. of bad tangent edges analytic = 0
; no. of G1 bad tangent edges analytic = 0
; no. of bad tangent edges uv_uv = 0
; no. of bad tangent edges boundary uv_uv = 0
; no. of bad tangent edges uv_nonuv = 0
; no. of bad tangent edges nonuv_nonuv = 0
; no. of bad tangent edges 3_4_sided = 0
; no. of surfaces = 4
; no. of discontinuous surfaces = 0
; percentage of good geom = 92
;()
;; 92
(hh:show-vertices-not-on-faces body1)
;; ()
; OUTPUT Result
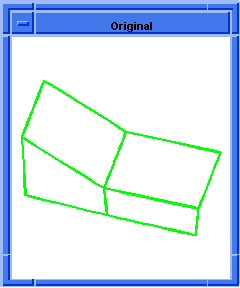  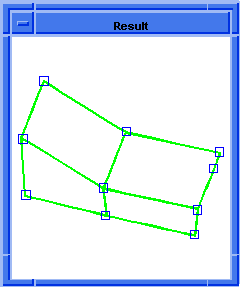
Figure. hh:show-vertices-not-on-faces |
[Top]
hh:simp-options
- Action
- Returns an object of Simp_Options.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- None
Syntax- (hh:simp-options "name-of-option" {value(s)} {options})
Arg Types-
Returns- Simp_Options
Description
This extension returns a Simp_Options object
for use in hh:analyze-simplify
and hh:simplify. This allows you
to set the simplification tolerance, maximum permissible circle radius, and the option
of switching curve and face simplification ON/OFF.
- Arguments
-
"name-of-option" specifies the name of the option.
-
value specifies the value of option.
-
options specifies the different Simp_Options.
- Note: Simp_Options object sets the parameters which govern the functionality of
hh:simplify and hh:simplify-analyze. The parameters are:
-
simplify_pos_tol has a default value of -1.
-
max_radius has a default value of -1.
-
do_curve_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1.
-
do_surface_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1.
- The default value of -1 for these parameters allows the respective algorithm
to undergo its default behavior.
- Errors
-
simplify_pos_tol option requires a real number as argument.
max_radius option requires a real number as argument.
do_curve_simplificaition option requires integer argument.
do_surface_simplification option requires integer argument.
Incorrect simplify option.
;1. Load the body
(part:load "xxx.sat" )
(define body (hh:combine (part:entities)))
;2. Make the simplification options object "Simp_Options"
(define so (hh:simp-options "do_curve_simplification" 1 "simplify_pos_tol" 0.001))
; To check the values of parameters of simp-options, type the option's name at the prompt
so
;3. For updating the parameters of an existing Simp_Options object, the same scheme command
; (hh:simp-options ) can be used by passing the options object as the last argument.
; (For example: hh:simp-options "do_surface_simplification" 1 so)
;4. Call simplification
(hh:simplify body so) |
[Top]
hh:simplify
- Action
- Simplifies spline geometry of the input body where possible.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_simplify_auto
Syntax- (hh:simplify body [simp-options] [acis-opts])
Arg Types-
Returns- body
Description
This extension replaces the spline surfaces
with the equivalent analytic representation where possible. This extension must follow the analysis
extension.
- Arguments
- body specifies the input body whose spline geometry is to be simplified.
simp-opts specifies the optional argument,
Simp_Options object that has been added since ACIS version R11. It sets the parameters
which govern the functionality of hh:simplify. The constructor of Simp_Options
sets the values of all the parameters as -1. The parameters that can be set are:
simplify_pos_tol has a default value of -1, that is the scheme command
computes a default value based on its own heuristics.-
max_radius has a default value of -1, that is, a maximum radius of 10,000.
-
do_curve_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1,
that is, curve simplication behavior is set to ON.
-
do_surface_simplification has values of -1, 0 (OFF), and 1 (ON). The default value is -1,
that is, surface simplication behavior is set to ON.
-
- Note:
(1) Selecting -1 as the value for any of these parameters enables the respective algorithm
to employ its default behavior.
(2) In ACIS R11 and later, both curve simplification and surface simplification are done
by default. In R10 and earlier, only surface simplification was done by default and there
was no curve simplification. To make hh:simplify behave as in R10 or an earlier
release, the caller of the extension requires creating a simp_options options object.
Do this by setting do_curve_simplification to "0" and passing this object into the scheme command.
- acis-opts is an optional argument for enabling journaling and versioning options.
Limitations- Entity must be a body. Individual sheet bodies or faces can be combined into a
single body using the hh:combine command.
; hh:simplify
; Replace spline surfaces with analytic surfaces
; where possible.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
; Make the simplification options object "Simp_Options"
(define so (hh:simp-options "do_curve_simplification" 1 "do_surface_simplification" 1 "simplify_pos_tol" 0.0001))
; To check the values of parameters of Simp_Options just type the option's name on the prompt
so
(hh:simplify body1 so)
; GEOM SIMPLIFICATION STATS:
; simplification tol= 0.0001
; no. of initial splines = 4
; no. of final splines = 4
; no. of planes made = 0
; no. of cylinders made = 0
; no. of spheres made = 0
; no. of tori made = 0
; no. of cones made = 0
;; #[entity 6 1] |
[Top]
hh:solve-analytic
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_analytic_auto
Syntax- (hh:solve-analytic body [acis-opts])
Arg Types-
Returns- body
Description
The analytic solver subphase attempts to heal all edges and vertices shared by analytic
surfaces.
- Arguments
- Input argument is the body on which the analyze and calculate stages of analytic solver
subphase of geometric building are to be executed.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:solve-analytic
; Load a file containing a bad part.
(define load (part:load 'heal1.sat))
;; load
; zoom the view.
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body.
(define body1 (hh:combine (part:entities)))
;; body1
; heal body
(define solve (hh:solve-analytic body1))
;; solve |
[Top]
hh:solve-gen-spline
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_gen_spline_auto
Syntax- (hh:solve-gen-spline body [acis-opts])
Arg Types-
Returns- body
Description
The generic spline solver attempts to heal generic tangential spline junction (for example,
the intersection curve is not an isoparametric curve of both splines in the intersection
curve).
- Arguments
- Input argument is the body on which the analyze and calculate stages of generic spline
solver subphase of geometric building are to be executed.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:solve-gen-spline
; Load a file containing a bad part.
(define load (part:load 'heal1.sat))
;; load
; zoom the view.
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body.
(define body1 (hh:combine (part:entities)))
;; body1
; heal body
(hh:solve-gen-spline body1))
;; #[entity 6 1] |
[Top]
hh:solve-isospline
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_isospline_auto
Syntax- (hh:solve-isospline body [acis-opts])
Arg Types-
Returns- body
Description
The isospline solver attempts to heal all edges shared by tangential isoparametric
surfaces (for example, the intersection curve is an isoparametric curve of both splines in the
intersection).
- Arguments
- Input argument is the body on which the analyze and calculate stages of isospline solver
subphase of geometric building are to be executed.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:solve-isospline
; Load a file containing a bad part.
(define load (part:load 'heal1.sat))
;; load
; zoom the view.
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body.
(define body1 (hh:combine (part:entities)))
;; body1
; heal body
(hh:solve-isospline body1))
;; #[entity 6 1] |
[Top]
hh:solve-sharp-edge
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_sharp_edge_auto
Syntax- (hh:solve-sharp-edge body [acis-opts])
Arg Types-
Returns- body
Description
The sharp edge solver attempts to heal all edges and vertices that are shared by
surfaces that intersect sharply. This includes nontangential surface junctions.
- Arguments
- Input argument is the body on which the analyze and calculate stages of sharp edge
subphase of geometric building are to be executed.
- The optional argument acis-opts helps enable journaling and versioning options.
; hh:solve-sharp-edge
; Load a file containing a bad part.
(define load (part:load 'heal1.sat))
;; load
; zoom the view.
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body.
(define body1 (hh:combine (part:entities)))
;; body1
; heal body
(hh:solve-sharp-edge body1))
;; #[entity 6] |
[Top]
hh:stitch
- Action
- Stitches faces into a single lump body where possible.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_stitch_auto
Syntax- (hh:stitch body [acis-opts])
Arg Types-
Returns- body
Description
This extension attempts to stitch the input set of faces into a solid or single sheet
body.
A small stitch tolerance is selected and all faces that can be stitched together at
that tolerance are processed. Then the tolerance is increased and the remaining faces are
processed and stitched if possible. This process is continued until all faces are stitched
together or a maximum stitch tolerance is exceeded.
- Arguments
- Argument body inputs the set of faces or individual sheet bodies that need to be
stitched. Individual sheet bodies or faces can be combined into a single body using the
hh:combine extension.
- incr-only sets incremental stitching.
- The optional argument acis-opts helps enable journaling and versioning options.
Limitations- The entity must be a body. Individual sheet bodies or faces can be combined into
a single body using the hh:combine extension.
; hh:stitch
; Stitch faces into a single lump body if possible.
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
; Prepare the body for healing
(hh:preprocess body1)
;; #[entity 6 1]
(hh:stitch body1)
; STITCH RESULTS :
; ================
; min_tol = 1e-05
; max_tol = 1
; no. solid lumps made = 0
; no. sheet lumps made = 1
; no. unshared loops = 1
; no. unshared edges = 9
;; #[entity 6 1] |
[Top]
hh:tighten-gaps
- Action
- Tightens gaps in the BODY that are bigger than the supplied desired_gap_tightness value.
Filename- scm/scmext/heal/gap_tighten_scm.cpp
APIs- api_tighten_gaps
Syntax- (hh:tighten-gaps body desired_gap_tightness [acis-opts] )
Arg Types-
Returns- body
Description
This extension attempts
to tighten gaps in the BODY that are bigger than the supplied desired_gap_tightness value.
For more details refer to the documentation for api_tighten_gaps.
- Arguments
- body whose gaps are attempted to be tightened by the user-supplied desired_gap_tightness value.
- desired_gap_tightness denotes the gap tightness value that a user desires in the body.
For example, a user can set the desired desired_gap_tightness value based on the
manufacturability requirement of the downstream application.
- acis-opts can be used to enable journaling and versioning options.
; hh:tighten-gaps
; Tightens gaps in a BODY that are bigger than
; supplied desired_gap_tightness value.
;
; Set the gap tightness value that a user
; desires in the body
(define desired_gap_tightness 0.01)
;; desired_gap_tightness
;
; Load a file in which the user intends to have
; the gaps tightened within the user's
; desired_gap_tightness value.
(part:load "xxx.sat")
;; (#[entity 1 1])
;
; Define the user supplied entity whose gaps are
; to be tightened, as body.
(define body (list-ref (part:entities) 0))
;; body
;
; Call hh:tighten-gaps with desired_gap_tightness
; to close gaps in the user supplied body within
; user's desired_gap_tightness value.
(hh:tighten-gaps body desired_gap_tightness)
;; #[entity 1 1]
;
; Set the check_level to 30
(option:set 'check_level 30)
;; 20
;
; Check the body after tightening gaps
(entity:check body)
;; checked:
;; 1 lumps
;; 1 shells
;; 0 wires
;; 6 faces
;; 6 loops
;; 24 coedges
;; 12 edges
;; 8 vertices
;; () |
[Top]
hh:terminate-body-for-healing
- Action
- Removes any existing healing attributes and attaches new healing attributes.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_end_body_for_healing
Syntax- (hh:terminate-body-for-healing body [acis-opts])
Arg Types-
Returns- body
- Arguments
- Input argument is the body whose existing healing attributes are to be replaced.
- The optional argument acis-opts can be used to enable journaling and versioning options.
; hh:terminate-body-for-healing
; Attach healing attributes
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1] #[entity 4 1]
;; #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 1076700200]
; Combine the faces into a body
(define body1 (hh:combine (list (entity 2)
(entity 3) (entity 4) (entity 5))))
;; body1
(hh:autoheal body1)
; ... (autoheal results)
;; (#[entity 6 1] 70 100)
(hh:terminate-body-for-healing body1)
;; #[entity 6 1] |
[Top]
hh:wrapup
- Action
- Deprecated: This scheme extension has been deprecated because it calls a deprecated API.
Filename- scm/scmext/heal/scm_heal.cpp
APIs- api_hh_wrapup_auto
Syntax- (hh:wrapup body [acis-opts])
Arg Types-
Returns- body
- Arguments
- Input argument is the body whose existing healing attributes are to be replaced.
- The optional argument acis-opts can be used to enable journaling and versioning options.
; hh:wrapup
; Load a file containing a bad part
(part:load "heal1.sat")
;; (#[entity 2 1] #[entity 3 1]
;; #[entity 4 1] #[entity 5 1])
; Zoom the view in order to see the part
(zoom-all)
;; #[view 5183062]
; Combine the faces into a body
(define body1 (hh:combine (part:entities)))
;; body1
; Heal the Body
(hh:wrapup body1)
;; #[entity 6 1] |
[Top]
© 1989-2007 Spatial Corp., a Dassault Systèmes company. All rights reserved.

![]()

































